Deeper Genomic Insights with DNBSEQ Complete WGS
More coverage, more insight, and more information.
Better Coverage
Uncover unknown areas of the genome.
Deeper Insights
Greater insights without parental testing.
Actionable Information
Valuable data for genetic disease research.
Complete WGS Overview
The NGS Limitations
Next-generation sequencing (NGS) technologies have enabled comprehensive whole-genome sequencing (WGS) using short DNA fragments, sequencing with SBS technology, and alignment to a reference genome. This method generates a single consensus sequence without distinguishing between variants on homologous chromosomes. However, this approach has limitations.
For instance, when mutations are detected, it’s unclear whether two mutations occur within the same gene or are distributed across two separate genes.
Phased Sequencing
Phased sequencing addresses this limitation by separating the consensus sequence into two individual sequence strands that occur together in phase, identifying alleles on both the maternal and paternal chromosomes.
It can provide valuable information for genetic disease research, such as analyzing structural variants, measuring allele-specific expression, identifying variant linkages, and more.
Affordable Phased Sequencing
Acquiring genomic phasing information has been challenging and costly, requiring specialized long-read sequencing technology. However, with the DNBSEQ™ Complete WGS (cWGS) solution, researchers can now generate highly accurate data while reducing costs.
The solution combines the output of WGS data generated through a PCR-free method with the DNBSEQ Complete WGS Kit which labels long DNA fragments, essentially reconstructing the long fragment DNA. Both outputs are analyzed with the DNBSEQ Complete WGS Analysis Software to generate the phasing information.
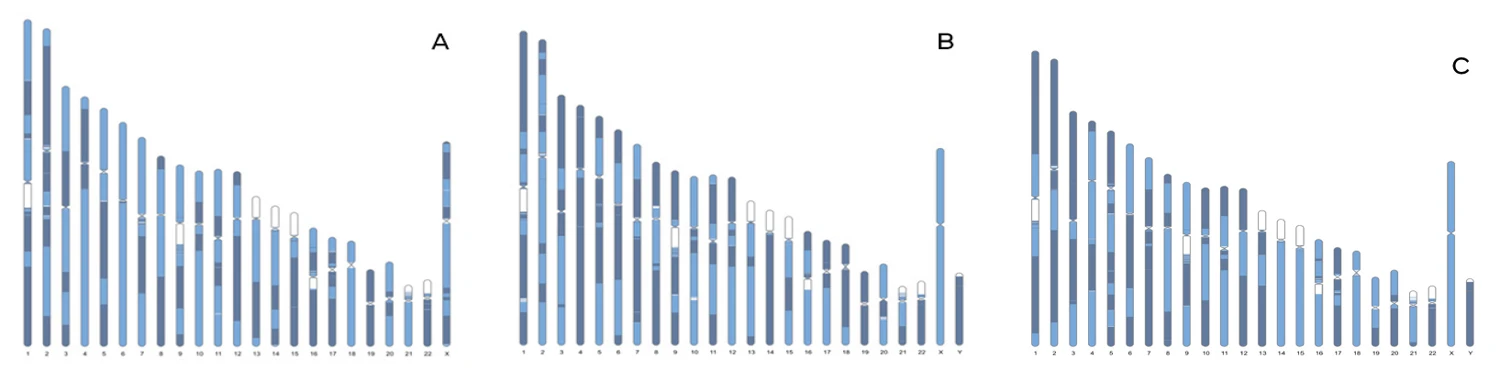
Visualizing Long-Phased Haplotype
Ideograms highlighting the long phased haplotype contigs a) H001 b) H002 and c) H005. DNA was isolated from approximately 20 million freshly harvested cells from GM12878 (HG001), GM24385 (HG002), and GM24631 (HG005) using the MGIEasy Magnetic Beads Genomic DNA Extraction Kit (1000010524). Isolated DNA was processed using the DNBSEQ Complete WGS kit using a single sample per reaction, followed by sequencing on the DNBSEQ-G400 sequencing instrument (PE100) to approximately 30X depth per sample. PCR-free libraries were generated using the MGIEasy FS PCR-Free DNA Library Prep Kit (1000013459) and sequenced to approximately 40X depth on the DNBSEQ-T7 sequencing instrument (PE150). FASTQ files were processed through the DNBSEQ Complete WGS Analysis Software.
| HG001 | HG002 | HG005 | |
|---|---|---|---|
| SNPs | 4,554,079 | 4,587,575 | 4,526,465 |
| Het SNPs | 2,298,495 | 2,309,109 | 2,100,359 |
| Indels | 1,281,296 | 1,305,204 | 1,298,396 |
| Het Indels | 621,814 | 622,817 | 579,681 |
| Barcode Split (%) | 81 | 83 | 82 |
| Long Fragments | 12,843,708 | 14,305,515 | 13,306,713 |
| Average Length of LFs | 82,290 | 71,055 | 86,510 |
| Phased het SNPs | 2,276,880 | 2,285,149 | 2,080,796 |
| Phased het Indels | 465,566 | 464,543 | 427,588 |
| Phaseblocks | 1,180 | 1,757 | 1,307 |
| Phaseblock N50 (mb) | 34.2 | 32.0 | 38.8 |
Multiplexed Sample Sequencing
DNBSEQ Complete WGS PanGenome data generated across a wide range of samples. DNA was isolated as previously described and processed in a similar manner, except that a multiplex of 10 samples per DNBSEQ Complete WGS reaction was performed. All sequencing was done on DNBSEQ-T7 using PE100 and PE150 for cWGS and PCR-free libraries, respectively. FASTQ files were processed through the DNBSEQ Complete WGS Analysis Software.
| GM19240 | GM20129 | HG00438 | HG00735 | HG01358 | HG01928 | HG02630 | HG03453 | HG03492 | |
|---|---|---|---|---|---|---|---|---|---|
| SNPs | 5,500,774 | 5,388,152 | 4,568,514 | 4,721,535 | 4,610,268 | 4,418,976 | 5,531,638 | 5,551,691 | 4,663,145 |
| Het SNPs | 3,099,775 | 3,107,351 | 2,184,486 | 2,522,639 | 2,319,823 | 1,896,402 | 3,107,115 | 3,146,205 | 2,372,570 |
| Indels | 1,588,814 | 1,564,934 | 1,374,318 | 1,389,518 | 1,391,288 | 1,375,203 | 1,646,903 | 1,600,805 | 1,450,998 |
| Het Indels | 809,848 | 807,535 | 593,246 | 668,840 | 621,973 | 523,835 | 810,262 | 820,508 | 635,577 |
| Barcode Split (%) | 85.62 | 73.94 | 82.87 | 80.09 | 85.68 | 79.22 | 79.75 | 84.70 | 81.35 |
| Long Fragments | 7,401,473 | 9,997,627 | 9,235,782 | 19,731,876 | 13,132,049 | 13,094,321 | 14,740,072 | 14,732,970 | 11,991,831 |
| Average Length of LFs | 111,019 | 94,982 | 93,695 | 52,037 | 67,054 | 79,207 | 79,052 | 58,517 | 88,550 |
| Phased het SNPs | 3,043,478 | 3,079,889 | 2,162,276 | 2,496,544 | 2,296,837 | 1,851,883 | 3,079,165 | 3,083,929 | 2,349,608 |
| Phased het Indels | 511,004 | 603,604 | 436,647 | 495,179 | 463,455 | 315,385 | 605,143 | 499,608 | 470,432 |
| Phaseblocks | 913 | 1,082 | 1,185 | 3,432 | 2,069 | 2,837 | 1,211 | 1,717 | 1,503 |
| Phaseblock N50 (mb) | 58.1 | 43.8 | 35.9 | 4.4 | 17.7 | 5.1 | 32.8 | 18.5 | 43.8 |
Complete WGS Workflow

Library Prep
SP-100 Automated Library Prep System
DNBSEQ FAST PCR-FREE FS Library Prep Set V2.0
DNBSEQ Complete WGS Set (96 Samples)
Complete WGS Data Analysis
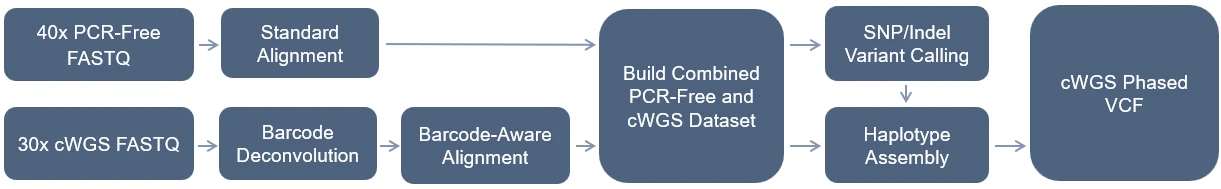
The DNBSEQ Complete WGS (cWGS) analysis pipeline enables the mapping, variant calling, and phasing of input FASTQ files from a PCR-free (PF) and a DNBSEQ Complete WGS (cWGS) library of the same sample. Running this pipeline results in a highly accurate and complete phased variant calling file (VCF). We recommend at least 40X coverage depth for the PCR-free library and 30X coverage depth for the cWGS library. Below is a visual summary of the pipeline.
DNBSEQ Complete WGS (cWGS) Analysis Pipeline
System Requirements for cWGS Analysis Pipeline
| Hardware Software | Software |
|---|---|
| Multicore computer (48 CPUs or more) | Linux CentOS 7 or later |
| Minimum 72 GB RAM | Root access may be necessary to install Singularity on the system |
| Storage may vary on on sample count and coverage; expect approximately 1 TB per sample | 14 hours per sample (with minimum system requirements) Per sample run time can be reduced by batching samples and utilizing additional CPUs |
Analysis Run Time
The expected analysis run time is 14 hours per sample, with the system meeting the minimum requirements. Per sample analysis, run time can be greatly reduced by batching samples and utilizing additional CPUs.
Specifications
| Methods | PCR-FREE | Complete WGS |
|---|---|---|
| Applications | Haplotype identification, structural variant identification, de novo | |
| Compatible Species | Human, simple plants (rice and lettuce), animals (dogs, moths, fish) | |
| Sample Type | DNA | |
| Input Quantity | 250 – 400 ng | 10 ng |
| Manual Assay Time | 3.2 hrs | 18 hrs |
| Run Time on SP-100* | ~ 2 hrs | ~ 9 hrs |
| Barcode | 48 unique barcodes | |
| Sequencer | DNBSEQ-T7 | |
| Read Length | PE150 | PE100 |

Ordering Information
| Category | Product | Cat. No. |
|---|---|---|
| Instrument | DNBSEQ-T7 Genetic Sequencer | 900-000698-00 |
| Automation | SP-100 Automated Library Prep System | 900-000206-00 |
| SP-Smart 8 Sample Preparation System | 900-000495-00 | |
| Sample Prep | MGIEasy Magnetic Beads Genomic DNA Extraction Kit | 1000010524 |
| Library Prep | DNBSEQ Fast PCR-FREE FS Library Prep Set V2.0 | 940-001314-00 |
| DNBSEQ Complete WGS (96 Samples) | 940-002564-00 | |
| Sequencing Reagents | DNBSEQ-T7 High-throughput Sequencing Set (DNBSEQ Complete WGS FCL PE150) | 940-002496-00 |
| DNBSEQ-T7 High-throughput Sequencing Set (FCL PE100) V1.0 | 940-000836-00 | |
| Hardware | MegaBOLT Bioinformatics Accelerator (Workstation Server) | 900-000677-00 |
| MegaBOLT Dongle | 058-000019-00 |
A Complete Solution for Every Step of Your Workflow

Kits & Reagents
No matter your workflow, our kits are simple to use and easily integrated. We also offer conversion kits for 3rd party library prep.

Lab Automation
With solutions from sample extraction to lab automation you can spend less time overseeing equipment and pursue more useful activities.





