Single-cell RNA Sequencing
Unprecedented insights into cellular diversity and function.
What is Single-cell RNA Sequencing?
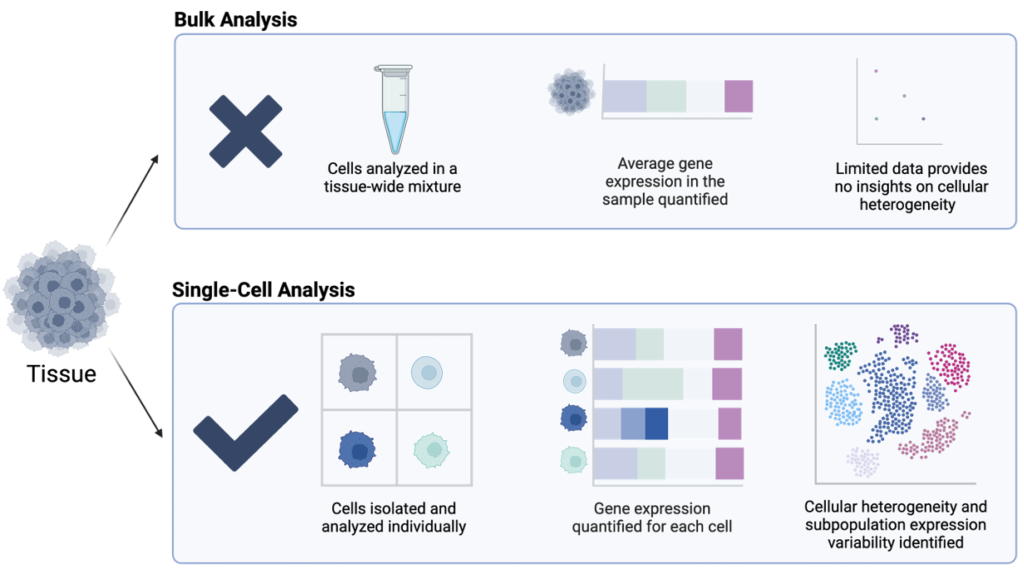
Single-cell sequencing is a cutting-edge technology that allows researchers to analyze individual cells’ genetic and molecular characteristics, providing unprecedented insights into cellular diversity and function.
Unlike traditional bulk sequencing, which averages signals from thousands of cells and masks the differences between individual cells, single-cell sequencing reveals the heterogeneity within a population, enabling researchers to identify distinct cell subpopulations, understand cell-specific gene expression patterns, and observe how individual cells contribute to overall tissue function or disease progression.
Applications of Single-cell RNA Sequencing
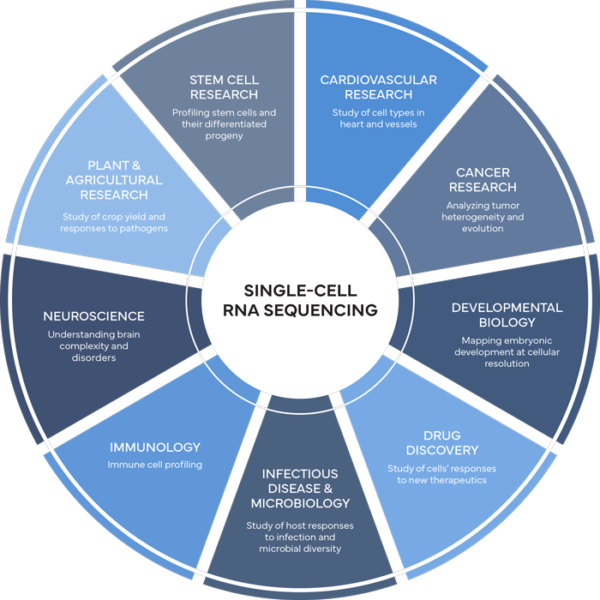
The field of single-cell genomics is advancing rapidly, providing new insights into complex biological systems—from the genomics of human cancer to the diversity of microbial ecosystems.
Common applications of single-cell sequencing include cancer research, cardiovascular research, developmental biology, drug discovery, infectious disease, immunology, neuroscience, Plant and Agricultural Research ,and stem cell research.
Single-cell Sequencing Workflow with DNBSEQ™ Technology
DNBSEQ sequencing technology is compatible with a range of single-cell library preparation and analysis solutions, including those from 10x Genomics, Parse, and Fluent Biosciences. Unlike traditional PCR amplification, DNBSEQ uses rolling circle replication (RCR), which ensures highly accurate sequencing with exceptional single-base resolution. This advanced approach eliminates common issues such as index hopping and reduces duplicate levels, leading to more reliable and precise results.
Single-cell sequencing can be cost-intensive due to the large number of cells that need to be sequenced. The DNBSEQ-G400 and DNBSEQ-T7 sequencing platforms address this challenge by offering higher accuracy, exceptional flexibility, and significantly improved cost efficiency, making them ideal solutions for single-cell applications.
Sample to Analysis Workflow

Sample Prep
Parse Biosciences:
Evercode Cell
Fixation v2 Kit

Single-cell Library Prep
10x Genomics:
Chromium Single Cell
Parse Biosciences:
Evercode WT Mini
Evercode WT
Evercode WT Mega
Fluent Biosciences:
PIPseqV

Library Conversion
DNBSEQ OneStep Library Conversion Kit

Analysis
Parse Biosciences:
Data Analysis Software
DNBSEQ-T7 with 10x Genomics: A Robust Alternative for High-Throughput Single-cell Sequencing Solution
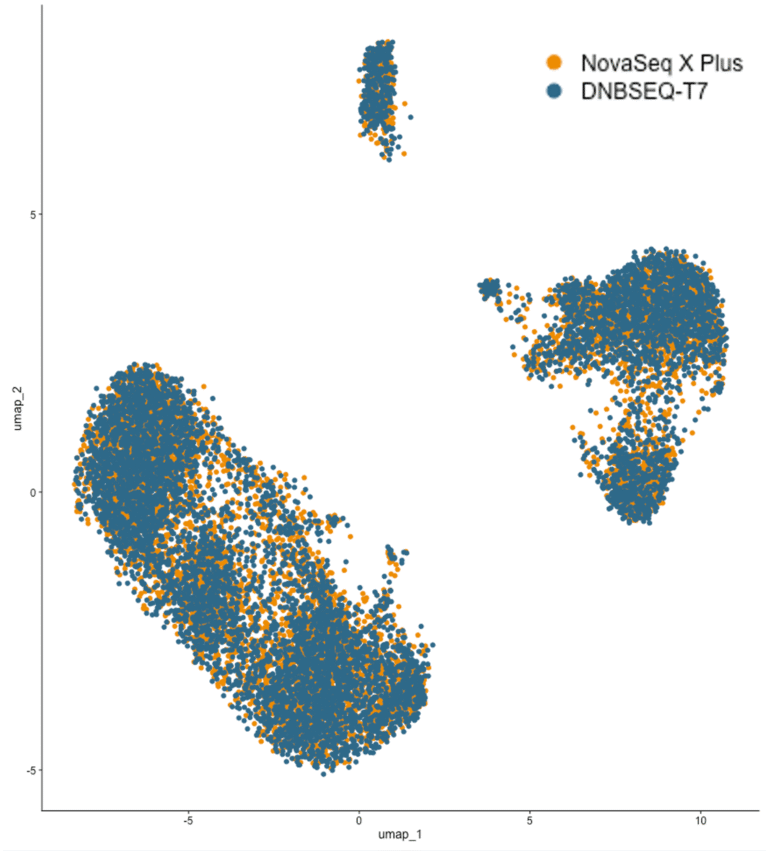
This study demonstrates a high-quality single-cell RNA-seq workflow using the 10x Genomics 3’ Kit v3.1 with Complete Genomics DNBSEQ-T7, benchmarked against Illumina NovaSeq X Plus. Using the same human lymphocyte sample, DNBSEQ-T7 showed comparable or slightly improved performance in mapping rates, Q30 scores, and cell-type resolution. It also captured more intronic and exonic reads, aiding detection of unspliced transcripts.
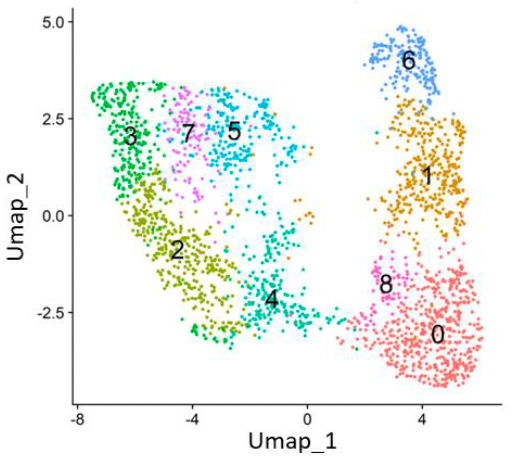
UMAP analysis confirmed its ability to clearly resolve diverse cell populations, validating DNBSEQ-T7 as a reliable, cost-effective platform for scRNA-seq.
High-Quality, Cost-Efficient Single-cell Sequencing Solution with Parse and DNBSEQ-T7
The DNBSEQ-T7 platform demonstrated comparable or slightly superior performance, with higher Q30 scores across barcodes, a higher median number of genes per cell, and lower mitochondrial and ribosomal gene content. Notably, unsupervised clustering resolved an NK cell cluster in the DNBSEQ-T7 dataset without the need for subclustering, highlighting its ability to capture subtle transcriptomic differences.
These results underscore the DNBSEQ-T7 as a powerful, high-throughput option for scRNA-seq with enhanced cost efficiency.
Single-cell Sequencing Solution by Fluent PIPseq and DNBSEQ-G400
The DNBSEQ-G400 sequencing platform is an excellent choice for routine single-cell RNA-seq analysis using the Fluent Biosciences PIPseq kit. With high sequencing quality (>93% Q30), full compatibility with the PIPseeker analysis pipeline, and performance that matches or exceeds the NextSeq 2000, the DNBSEQ-G400 delivers reliable and precise results. Moreover, the cost of generating PIPseq data with this system is estimated at just $0.11 per cell, offering an affordable solution for single-cell studies.

Decoding Cellular Diversity at Single-cell Resolution
Researchers are utilizing DNBSEQ technology to perform high-resolution, full-length single-cell RNA sequencing, thereby revealing the transcriptional complexity within and across various cell types. These insights are critical for understanding gene regulation, disease mechanisms, and cellular responses in health and disease. By combining deep transcript coverage with scalable throughput, this approach unlocks new potential for applications ranging from immunology to cancer biology, while remaining cost-effective and accessible.
Learn how Dr. Christoph Ziegenhain, an assistant professor at the Karolinska Institute & CEO of Xpress Genomics, is studying transcriptional bursting and allele-specific expression in scRNA-seq datasets obtained from DNBSEQ sequencing platforms.


