The Race to Affordable Next-Generation Sequencing Costs: From the $1,000 Genome to the Sub-$100 Genome
Sep 16, 2024
Since the beginning of the Next-Generation Sequencing (NGS) era, one chart has graced industry slide decks more than any other, illustrating the significant drop in Next-Generation Sequencing costs.
If you’ve set foot inside a genomics conference or half-watched a webinar about a new sequencing technology, then surely you have seen this figure.
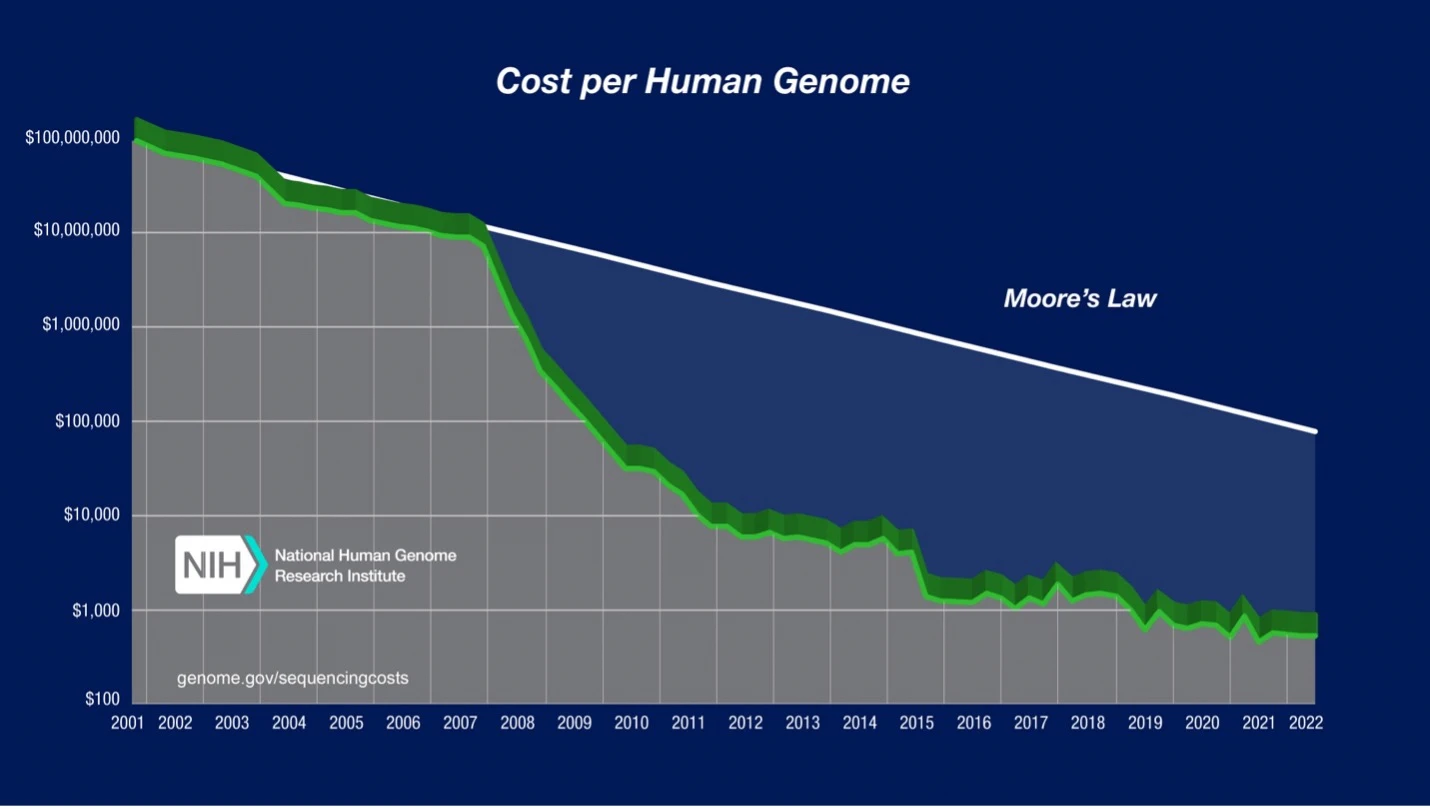
Published by the National Human Genome Research Institute (NHGRI), the chart depicts the dramatic reduction in per-genome sequencing costs from the early 2000’s until 2022, when the numbers were last updated. Notice the log-scaled vertical axis. We can see that the cost of sequencing a human genome came down a whopping five OOMs (orders of magnitude) within the span of about 20 years.
The most precipitous drop in Next-Generation Sequencing costs (~three OOMs) occurs between 2007 and 2011—that’s when modern high-throughput (or “next-generation”) methods began to supplant Sanger sequencing.
Prior to 2007, Next-Generation Sequencing costs largely tracked with a kind of ‘Moore’s Law’ for genomics, a trendline which saw sequencing cost-per-base halving roughly every 2 years. (Note: Moore’s Law specifically refers to the observation that the # of transistors you can fit on a microprocessor has doubled every 2 years since the late Sixties, so its application to genomics is a bit of a misnomer, though a fitting one.) In the years after 2007, sequencing efficiency has outpaced Moore’s Law.
It’s difficult to overstate what a sea change this was. Suddenly, techniques such as whole-genome sequencing and exome sequencing were inexpensive enough that they could be used to inform treatments for individual patients. The era of personalized genomics had begun. Genomicists set their sights on the “$1,000 genome,” a price point at which genome sequencing would become accessible for widespread clinical and research use.
Complete Genomics emerged as a formidable competitor in the race for the $1,000 genome. In 2009, Helicos Biosciences made waves with their claim that they had sequenced a human genome for a cost in consumables of just $48,000. That same year, Complete Genomics launched their whole-genome sequencing service, charging 1/10th as much: as little as $5,000/genome for bulk orders.
Then, in Q1 of 2014, Illumina released the HiSeq X Ten, an ultra-high throughput sequencer that achieved the $1,000/genome benchmark via both technological innovations and a favorable economy of scale. Illumina may have been first past the post, but the race was still on.
Next-Generation Sequencing Costs: The Race for the $100 Genome
Though momentous, the $1,000 per genome price point was not a cure-all. For starters, when we talk about the “$1,000 Genome,” we mean that the per-genome cost of sequencing reagents is $1,000. Although sequencing reagents take up the largest slice of the pie, there are other substantial Next-Generation Sequencing costs on the path from sample to sequenced genome, including labor, administration, library preparation, analysis, data storage, and amortized instrument cost.
As a case in point, consider the NHGRI dataset shown in the graph above. Their analysis considers a broad array of costs in addition to sequencing reagents, and because of that, the “real” per-genome cost does not drop below $1,000 until 2019—five years after humanity had purportedly achieved the “$1,000 Genome.”
Consequentially, the sequencing industry set its sights on a new goal: the $100 genome. The $100 genome would enable a “sequence everything” approach, where the cost would be low enough to justify sequencing research and clinical samples that would have gone un-sequenced in years prior.
As an oft-cited example, consider target capture. Hybridization-capture workflows are laborious and costly. As Next-Generation Sequencing costs and analysis costs continue to plummet, we can envision a future where target capture goes extinct because it’s cheaper to just sequence all the DNA or RNA in a given sample and deal with it during analysis.
Things start to get interesting in 2023. At AGBT 2023, Complete Genomics releases the DNBSEQ-T20x2 sequencing platform, an ultra-high throughput sequencer designed for population genomics projects. This new instrument could produce a whopping 50,000 WGS per year for under $100 per 30x human genome. If Illumina had won the race to the $1,000 genome, then Complete Genomics clearly had won the race to the sub-$100 genome.
Around the same time, Illumina announced the release of the NovaSeq X plus in 2024, claiming they had achieved a $200 genome with 25B flow cell. Shortly after, Complete Genomics announces that our flagship high-throughput sequencer DNBSEQ-T7 can sequence a $150 genome and that’s just the starting point. Finally, at AGBT 2024, new entrant Ultima Genomics announces that their UG100 can attain a $100 genome.
High Throughput Sequencer Shootout (Q3 2024)
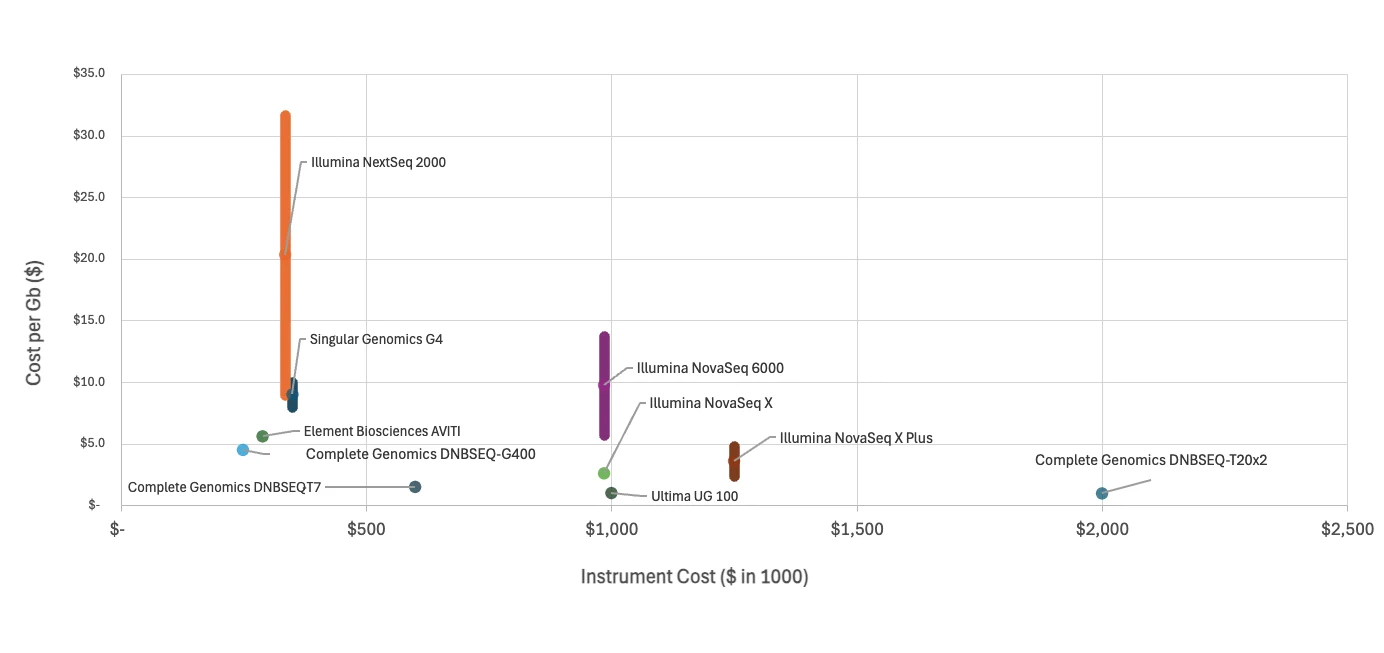
With so many sequencers on the scene, how does one decide which to purchase? For many organizations, the main factor is cost: which sequencer will be least expensive to purchase and operate at a given throughput?
First, let’s take a look at “mid throughput” sequencers—those which generate ~1,000 Gb per sequencing run. These include the DNBSEQ-G400 from Complete Genomics, as well as Element’s AVITI, Singular Genomics’ G4, and Illumina’s NextSeq 2000. The DNBSEQ-G400 emerges as the clear winner in the mid-throughput category, boasting both a lower instrument cost ($249k) and a lower cost to operate ($450 per genome) than its competitors.
Next, let’s consider “high throughput” sequencers—those which generate ~10,000 Gb per sequencing run. While “ultra-high throughput” sequencers do exist, such as the aforementioned DNBSEQ-T20x2 sequencing platform, they serve specialized applications (such as population genomics), and they have not experienced the same market adoptions as their “high throughput” counterparts have. For a wide swath of prospective buyers, high throughput sequencers exist in a sweet spot where the initial investment justifies the per-base cost savings.
Complete Genomics’ DNBSEQ-T7 handily beats all of Illumina’s offerings in both instrument cost and cost to operate. In fact, Illumina’s flagship NovaSeq X plus instrument costs over twice as much as the DNBSEQ-T7, and the cost per gb is substantially greater, at $200 per genome compared to the DNBSEQ-T7’s $150 per genome.
Next, let’s consider the UG100. At $100 per genome, the UG100 edges out the T7’s $150 per genome, but you’ll pay a pretty penny for it—the UG100 instrument costs 2.5X as much as the T7. And even if you can justify that cost, realize that you’ll be taking on a substantial risk: Ultima Genomics’ instruments and chemistry are new to the market, so they’re not field-tested. Complete Genomics’ DNBSEQ technology has proven reliability in the field.
Among high-throughput sequencers, the Complete Genomics DNBSEQ-T7 stands out as the most compelling option, offering a balanced combination of lower initial investment, reduced operational costs, and flexible throughput. For organizations seeking to maximize return on investment, the DNBSEQ-T7 is the most reliable and cost-effective choice in today’s market landscape.
Subscribe for exclusive content and expert insights.









