DNBSEQ-T1+ Sequencer
From Discovery
to Production
All-in-One Sequencer
DNBSEQ-T1+ Sequencer
From Discovery to Production
All-in-One Sequencer
DNBSEQ-T1+ Sequencer
DNBSEQ-T1+, powered by DNBSEQ™ technology, is a highly efficient mid-throughput sequencer with Q40 accuracy, delivering 25Gb – 1.2Tb of data in under 24 hours. Its optimized performance and flexible sequencing options enable scientists to seamlessly explore genomics—from discovery to validation and production-scale sequencing—while maintaining cost-effectiveness.
Speed
Unmatched performance, generating 1.2 Tb of data in ~ 22.5 hours
Discovery
Enhance breakthroughs with Q40 accuracy and flexible read options
Possibilities
Unlock multi-omics potential with versatile, cost-effective sequencing options
See the DNBSEQ-T1+ in Action
DNBSEQ-T1+ Sequencer
DNBSEQ-T1+, powered by DNBSEQ™ technology, is a highly efficient mid-throughput sequencer with Q40 accuracy, delivering 25Gb – 1.2Tb of data in under 24 hours.
Its optimized performance and flexible sequencing options enable scientists to seamlessly explore genomics—from discovery to validation and production-scale sequencing—while maintaining cost-effectiveness.
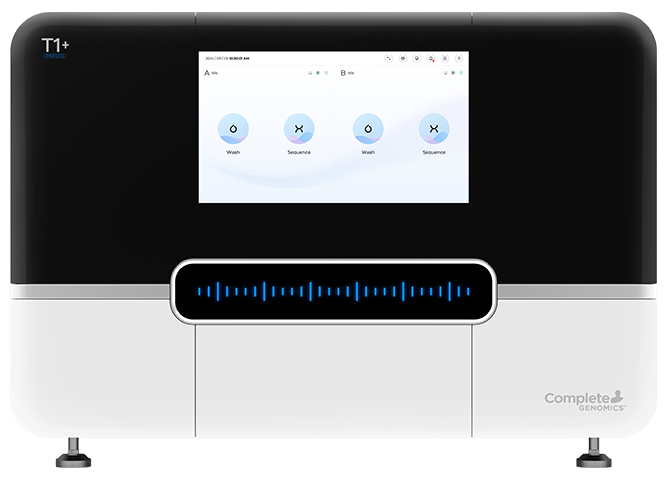
Seamless Sample to Analysis Workflow
DNBSEQ-T1+ streamlines the DNB preparation and loading process with onboard “DNB Make and Load” technology, enabling a one-click, quantification-free workflow for these steps.
The advanced DNBSEQ-T1+ ARS model further enhances efficiency by offering onboard secondary data analysis, supporting Whole Genome Sequencing (WGS), Whole Exome Sequencing (WES), and RNA Sequencing (RNA-seq).

Unrivaled Flexibility
DNBSEQ-T1+ offers independent dual flow
cell operation, allowing researchers to run two flow
cells independently, at any time, with various sequencing
options and flow cell types.
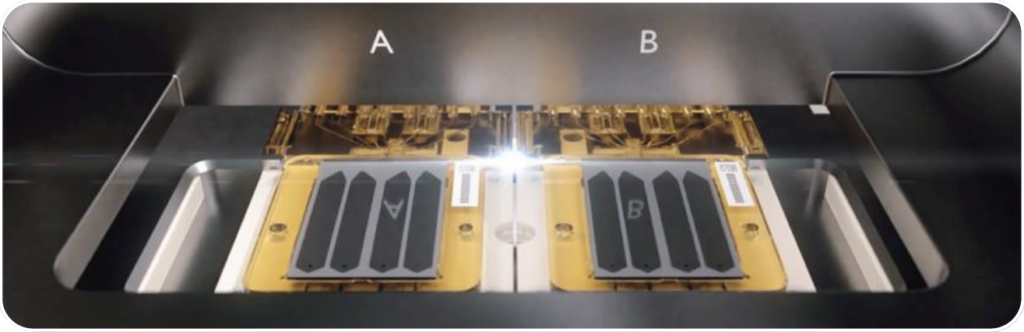
Three Flow Cells Specifications
There are three types of flow cells designed to meet diverse needs (refer to the performance parameter table). Each flow cell features multiple individually addressable lanes, allowing researchers to:
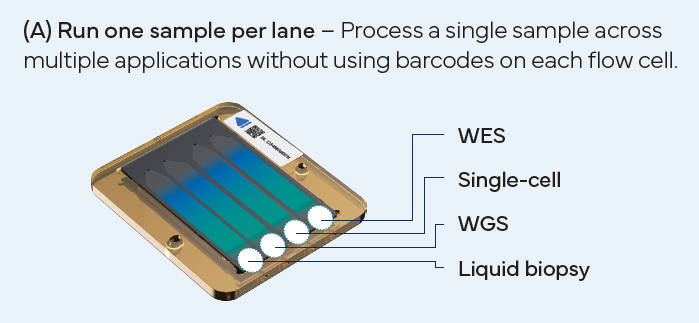
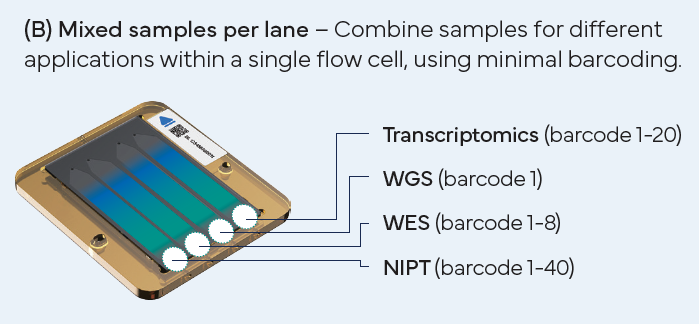
Versatile Sequencing Options
DNBSEQ-T1+ supports up to six sequencing read lengths (SE50/100 and PE50/100/150/300) across three flow cell types, enabling an exceptionally wide range of applications, including WGS, WES, Single-cell, Spatial Transcriptomics, Infectious Disease, and more.
Performance Parameters
| Flow Cell Type | Readsa | Read Length | Data Outputb | Run Timec | Q30 | Q40 |
|---|---|---|---|---|---|---|
FCS | 2 lanes | 500 M | SE100 | 50 Gb | 8 hr | >93% | >90% |
| PE150 | 150 Gb | 20.5 hr | >93% | >90% | ||
| PE300 | 300 Gb | 36 hr | >85% | >75% |
| Flow Cell Type | Readsa | Read Length | Data Outputb | Run Timec | Q30 | Q40 |
|---|---|---|---|---|---|---|
FCM | 2 lanes | 1000 M | SE100 | 100 Gb | 9 hr | >93% | >90% |
| PE150 | 300 Gb | 21.5 hr | >93% | >90% |
| Flow Cell Type | Readsa | Read Length | Data Outputb | Run Timec | Q30 | Q40 |
|---|---|---|---|---|---|---|
FCL | 4 lanes | 2000 M | SE100 | 200 Gb | 10 hr | >93% | >90% |
| PE150 | 600 Gb | 23 hr | >93% | >90% |
a. The maximum number of effective reads are based on the sequencing of an internal standard library. Actual output may vary with sample types and library preparation methods.
b. The listed data output is per flow cell, meaning the total output per run doubles when utilizing both flow cells.
c. Run time is based on the dual flow cell module, including sample loading, sequencing, and FASTQ generation.
FCS and FCL will be available in H2 2025, while FCS PE300 and FCM flow cells will be available in 2026.
