What is Spatial Transcriptomics? Transforming Gene Expression Analysis
Sep 19, 2024
Discover how spatially resolved transcriptomes reveal cellular and tissue function and provide a complete view of biological systems.
Revealing the Spatial Dimension of Biology
Spatial transcriptomics is revolutionizing our understanding of gene expression by allowing researchers to measure and visualize gene activity within the context of tissue architecture.
For a deeper dive into its applications, visit our dedicated page on Spatial Transcriptomics.
This technology combines the power of next-generation sequencing with spatial resolution, providing unprecedented insights into cellular organization, function, and interactions within complex tissues.
The recognition of spatially resolved transcriptomics as the “Method of the Year 2020” by Nature Methods underscores its transformative potential in advancing our understanding of biological systems.
This technology is already accelerating drug discovery, therapy development, and our comprehension of complex diseases by providing gene expression information in a spatial dimension.
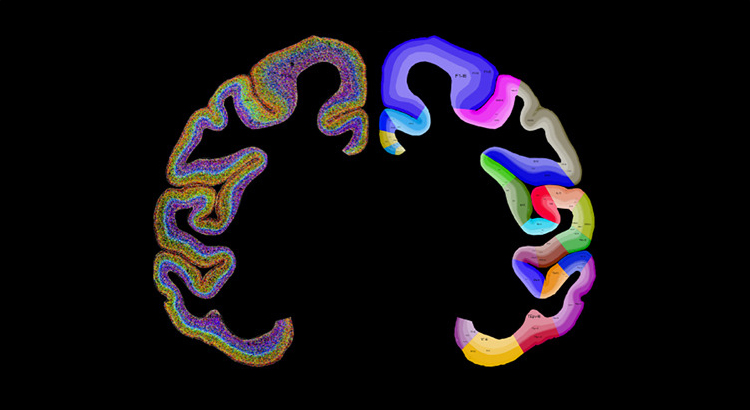
What is Spatial Transcriptomics?
Spatial transcriptomics provides vital biological context by coupling expression data with cell localization information within intact tissue samples.
By preserving spatial information, this approach allows researchers to map gene expression directly onto the tissue, providing insights into cellular interactions and the microenvironment that are not accessible with traditional methods.

Spatial transcriptomics offers three primary advantages over traditional transcriptomics:
- Retaining tissue context to preserve the spatial relationships between cells and their microenvironment.
- Revealing spatial gene expression patterns to uncover how gene expression varies across different regions of a tissue.
- Enabling the study of tissue heterogeneity to allow for the identification of distinct cell populations and their spatial organization.
There are multiple approaches to capturing spatially resolved transcriptomes.
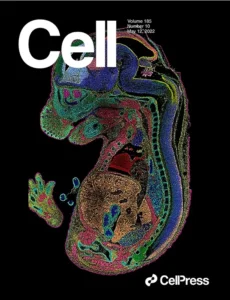
Our core technology, Spatio-Temporal Enhanced Resolution Omics sequencing (Stereo-seq), was pioneered by scientists at STOmics and featured on the cover of Cell in 2022. Stereo-seq offers unique benefits compared to other methodologies, including enabling a large field of view, providing single-cell resolution, and serving as a species-agnostic approach.
How is Spatial Transcriptomics Used?
Spatial transcriptomics is making significant impacts across various fields of study, from basic research into tissue structure to clinical research developing targeted therapeutics based on newly identified biomarkers.
Key areas of study, include:
Cancer Research
To examine tumor heterogeneity and the tumor microenvironments.
Tumors are often composed of a mix of different cell types with varying gene expression profiles. Spatial transcriptomics allows for the study of this heterogeneity, providing insights into tumor evolution and resistance to therapy.
Learn how spatial transcriptomics enhances cancer research on our applications page.
By mapping gene expression in the context of tissue architecture, spatial transcriptomics helps researchers understand the interactions between cancer cells and their surrounding environment. This can lead to the identification of new therapeutic targets and biomarkers for cancer treatment.
Scientists use a multi-omics approach to study spatial heterogeneity in ovarian cancer.
Learn more
Researchers detect a novel immunosuppressive invasion zone in liver cancer.
Learn more
Neuroscience
To investigate brain region-specific gene expression and neuronal subtypes.
The brain is a highly complex organ with diverse cell types organized in a specific spatial arrangement. Spatial transcriptomics enables gene expression mapping across different brain regions, contributing to our understanding of brain function and neurodevelopmental disorders.
In diseases like Alzheimer’s and Parkinson’s, spatial transcriptomics can be used to study changes in gene expression within affected brain regions, offering clues to disease mechanisms and potential treatment strategies.
Researchers generate comprehensive cell type taxonomy of the macaque cerebral cortex.
Learn more
Developmental Biology
To map gene expression changes during embryogenesis and organ development.
During embryogenesis, cells differentiate and organize into complex structures. Spatial transcriptomics allows scientists to track these changes in gene expression over time, providing insights into the molecular processes that drive development.
Understanding how organs form and develop is crucial for regenerative medicine. Spatial transcriptomics can identify key regulatory genes involved in organogenesis, potentially paving the way for novel therapeutic approaches.
Scientists use Stereo-seq to understand axolotl brain regeneration.
Learn more
Application Note
Construction of a Spatiotemporal Transcriptomic Atlas of Zebrafish Embryogenesis
Learn how Stereo-seq technology was used to profile zebrafish embryo sections covering critical time points during the first 24 hours of development.

How is Spatial Transcriptomics Used?
Spatial transcriptomics uses three main principles to couple gene expression and spatial context effectively.
Preserving spatial information
Unlike traditional bulk RNA sequencing, spatial transcriptomics maintains the original position of transcripts within a tissue sample.
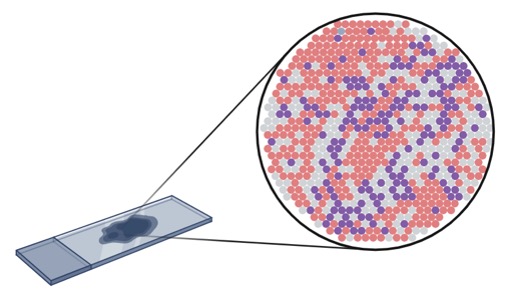
Capturing gene expression data
The technology quantifies mRNA molecules, providing a snapshot of gene activity across the tissue.
Combining histology with molecular profiling
Spatial transcriptomics integrates visual tissue morphology with high-throughput sequencing data.
Key Steps of Sequencing-based Spatial Transcriptomics
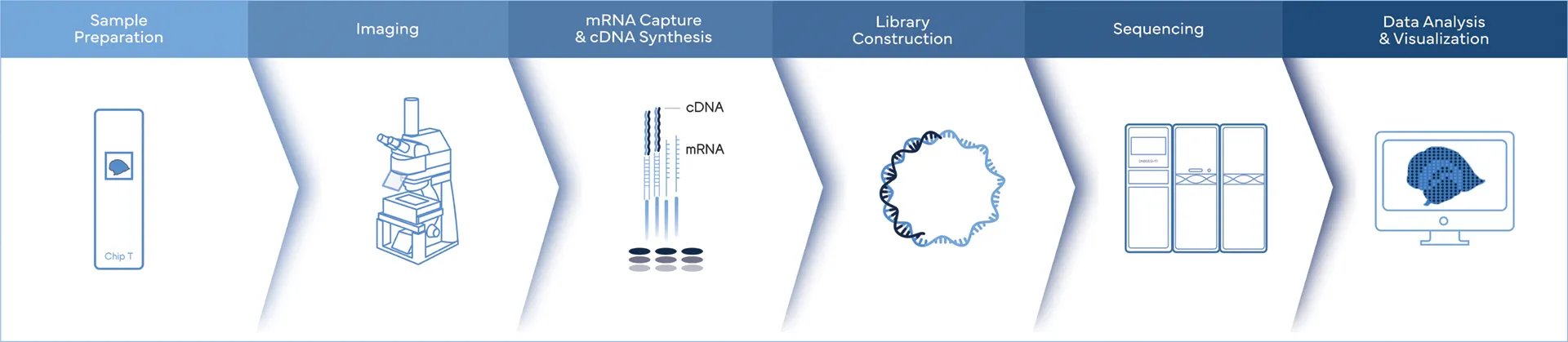
- Sample Preparation and Imaging: A tissue section is preserved on a specialized slide and visualized using microscopy.
- RNA Capture: The tissue is exposed to barcodes that bind to RNA molecules. These barcodes are spatially indexed, meaning each location on the slide has a unique identifier.
- Sequencing: The captured RNA is then sequenced, with the spatial barcodes allowing the sequence data to be mapped back to specific locations on the tissue.
- Data Analysis and Visualization: Bioinformatics tools reconstruct gene expression maps, allowing researchers to visualize where specific genes are active within the tissue.
What is Single Cell Spatial Transcriptomics?
Single-cell spatial transcriptomics is an advanced technique that analyzes mRNA expression profiles of individual cells while preserving their spatial context within intact tissue samples.
This method combines the high-resolution gene expression profiling of single-cell RNA sequencing with the spatial information traditionally lost in dissociation-based approaches.
The resolution of single-cell spatial transcriptomics can be remarkably high, with the best methods achieving subcellular resolution. This allows for precise mapping of gene expression patterns within individual cells and across tissue structures, enabling researchers to uncover complex spatial relationships in gene activity that were previously unobservable.
Spatial transcriptomics addresses key limitations of traditional single-cell RNA sequencing (scRNA-seq) methods. While scRNA-seq has provided novel insights into cellular heterogeneity, it requires the dissociation of tissues into single cells, destroying spatial context and making it challenging to study certain cell types, such as neurons in the brain.
What is Stereo-seq?
Stereo-seq is a cutting-edge spatial transcriptomics technology at the core of all Complete Genomics spatial biology solutions. This innovative approach enables researchers provides a powerful tool for understanding complex biological processes at the molecular level.
Subscribe for exclusive content and expert insights.